[ad_1]

Structure prediction is a elementary drawback in molecular science as a result of the construction of a molecule determines its properties and features. In current years, deep studying strategies have made outstanding progress and affect on predicting molecular constructions, particularly for protein molecules. Deep studying strategies, comparable to AlphaFold and RoseTTAFold, have achieved unprecedented accuracy in predicting essentially the most possible constructions for proteins from their amino acid sequences and have been hailed as a recreation changer in molecular science. However, this technique gives solely a single snapshot of a protein construction, and construction prediction can not inform the entire story of how a molecule works.
Proteins usually are not inflexible objects; they’re dynamic molecules that may undertake totally different constructions with particular chances at equilibrium. Identifying these constructions and their chances is important in understanding protein properties and features, how they work together with different proteins, and the statistical mechanics and thermodynamics of molecular techniques. Traditional strategies for acquiring these equilibrium distributions, comparable to molecular dynamics simulations or Monte Carlo sampling (which makes use of repeated random sampling from a distribution to realize numerical statistical outcomes), are sometimes computationally costly and should even turn into intractable for advanced molecules. Therefore, there’s a urgent want for novel computational approaches that may precisely and effectively predict the equilibrium distributions of molecular constructions from fundamental descriptors.
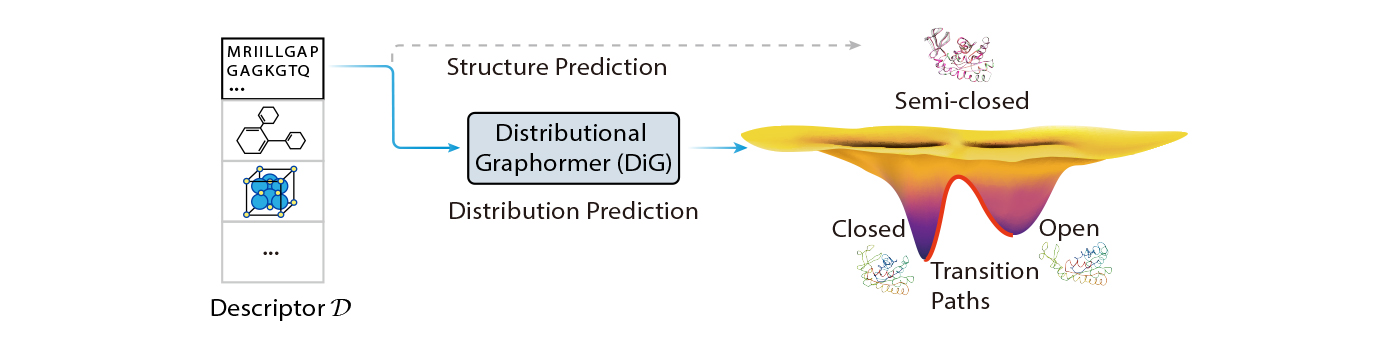
In this weblog publish, we introduce Distributional Graphormer (DiG), a brand new deep studying framework for predicting protein constructions in line with their equilibrium distribution. It goals to deal with this elementary problem and open new alternatives for molecular science. DiG is a big development from single construction prediction to construction ensemble modeling with equilibrium distributions. Its distribution prediction functionality bridges the hole between the microscopic constructions and the macroscopic properties of molecular techniques, that are ruled by statistical mechanics and thermodynamics. Nevertheless, it is a great problem, because it requires modeling advanced distributions in high-dimensional house to seize the chances of various molecular states.
DiG achieves a novel answer for distribution prediction by way of an development of our earlier work, Graphormer, which is a general-purpose graph transformer that may successfully mannequin molecular constructions. Graphormer has proven wonderful efficiency in molecular science analysis, demonstrated by functions in quantum chemistry and molecular dynamics simulations, as reported in our earlier weblog posts (see right here and right here for extra particulars). Now, we’ve superior Graphormer to create DiG, which has a brand new and highly effective functionality: utilizing deep neural networks to straight predict goal distribution from fundamental descriptors of molecules.
Spotlight: On-Demand EVENT
Microsoft Research Summit 2022
On-Demand
Watch now to find out about a few of the most urgent questions going through our analysis group and eavesdrop on conversations with 120+ researchers round how to make sure new applied sciences have the broadest attainable profit for humanity.
DiG tackles this difficult drawback. It relies on the thought of simulated annealing, a traditional technique in thermodynamics and optimization, which has additionally motivated the current improvement of diffusion fashions that achieved outstanding breakthroughs in AI-generated content material (AIGC). Simulated annealing produces a posh distribution by steadily refining a easy distribution by way of the simulation of an annealing course of, permitting it to discover and settle in essentially the most possible states. DiG mimics this course of in a deep studying framework for molecular techniques. AIGC fashions are sometimes based mostly on the thought of diffusion fashions, that are impressed by statistical mechanics and thermodynamics.
DiG can be based mostly on the thought of diffusion fashions, however we convey this concept again to thermodynamics analysis, making a closed loop of inspiration and innovation. We think about scientists sometime will be capable to use DiG like an AIGC mannequin for drawing, inputting a easy description, comparable to an amino acid sequence, after which utilizing DiG to rapidly generate lifelike and various protein constructions that comply with equilibrium distribution. This will tremendously improve scientists’ productiveness and creativity, enabling novel discoveries and functions in fields comparable to drug design, supplies science, and catalysis.
How does DiG work?
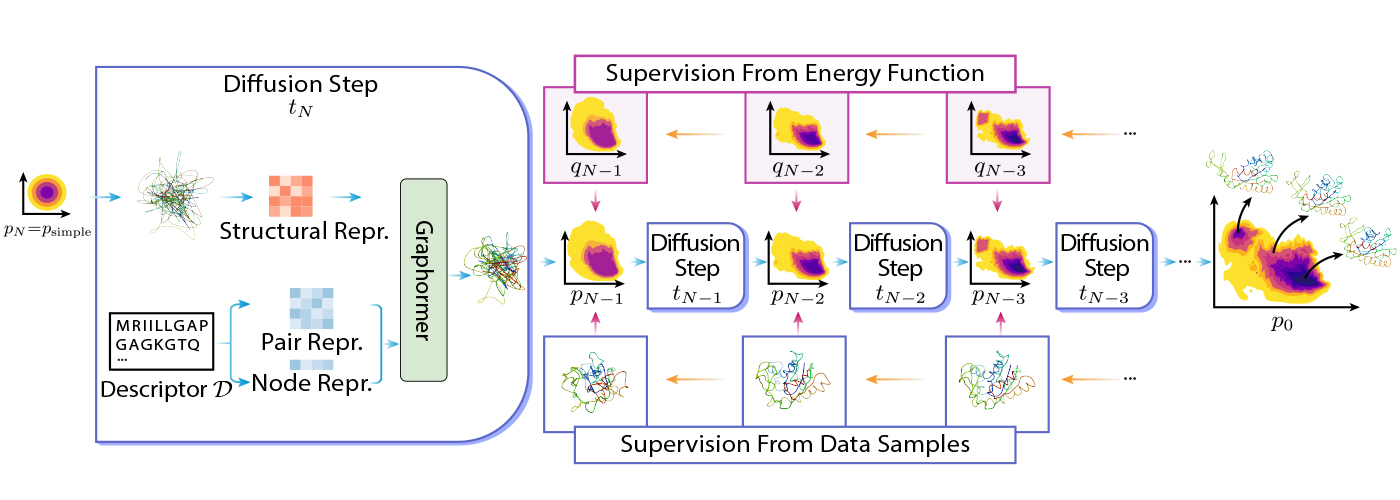
DiG relies on the thought of diffusion by remodeling a easy distribution to a posh distribution utilizing Graphormer. The easy distribution could be a customary Gaussian, and the advanced distribution might be the equilibrium distribution of molecular constructions. The transformation is completed step-by-step, the place the entire course of mimics the simulated annealing course of.
DiG might be educated utilizing several types of information or info. For instance, DiG can use power features of molecular techniques to information transformation, and it might probably additionally use simulated construction information, comparable to molecular dynamics trajectories, to study the distribution. More concretely, DiG can use power features of molecular techniques to information transformation by minimizing the discrepancy between the energy-based chances and the chances predicted by DiG. This strategy can leverage the prior information of the system and practice DiG with out stringent dependency on information. Alternatively, DiG can even use simulation information, comparable to molecular dynamics trajectories, to study the distribution by maximizing the probability of the info below the DiG mannequin.
DiG exhibits equally good generalizing talents on many molecular techniques in contrast with deep learning-based construction prediction strategies. This is as a result of DiG inherits some great benefits of superior deep-learning architectures like Graphormer and applies them to the brand new and difficult activity of distribution prediction. Once educated, DiG can generate molecular constructions by reversing the transformation course of, ranging from a easy distribution and making use of neural networks in reverse order. DiG can even present the likelihood estimation for every generated construction by computing the change of likelihood alongside the transformation course of. DiG is a versatile and normal framework that may deal with several types of molecular techniques and descriptors.
Results
We display DiG’s efficiency and potential by way of a number of molecular sampling duties protecting a broad vary of molecular techniques, comparable to proteins, protein-ligand complexes, and catalyst-adsorbate techniques. Our outcomes present that DiG not solely generates lifelike and various molecular constructions with excessive effectivity and low computational prices, however it additionally gives estimations of state densities, that are essential for computing macroscopic properties utilizing statistical mechanics. Accordingly, DiG presents a big development in statistically understanding microscopic molecules and predicting their macroscopic properties, creating many thrilling analysis alternatives in molecular science.
One main software of DiG is to pattern protein conformations, that are indispensable to understanding their properties and features. Proteins are dynamic molecules that may undertake various constructions with totally different chances at equilibrium, and these constructions are sometimes associated to their organic features and interactions with different molecules. However, predicting the equilibrium distribution of protein conformations is a long-standing and difficult drawback as a result of advanced and high-dimensional power panorama that governs likelihood distribution within the conformation house. In distinction to costly and inefficient molecular dynamics simulations or Monte Carlo sampling strategies, DiG generates various and functionally related protein constructions from amino acid sequences at a excessive pace and a considerably lowered price.
DiG can generate a number of conformations from the identical protein sequence. The left facet of Figure 3 exhibits DiG-generated constructions of the principle protease of SARS-CoV-2 virus in contrast with MD simulations and AlphaFold prediction outcomes. The contours (proven as strains) within the 2D house reveal three clusters sampled by intensive MD simulations. DiG generates extremely related constructions in clusters II and III, whereas constructions in cluster I are undersampled. In the fitting panel, DiG-generated constructions are aligned to experimental constructions for 4 proteins, every with two distinguishable conformations similar to distinctive practical states. In the higher left, the Adenylate kinase protein has open and closed states, each properly sampled by DiG. Similarly, for the drug transport protein LmrP, DiG additionally generates constructions for each states. Here, word that the closed state is experimentally decided (within the lower-right nook, with PDB ID 6t1z), whereas the opposite is the AlphaFold predicted mannequin that’s according to experimental information. In the case of human B-Raf kinase, the key structural distinction is localized within the A-loop area and a close-by helix, that are properly captured by DiG. The D-ribose binding protein has two separated domains, which might be packed into two distinct conformations. DiG completely generated the straight-up conformation, however it’s much less correct in predicting the twisted conformation. Nonetheless, moreover the straight-up conformation, DiG generated some conformations that seem like intermediate states.
Another software of DiG is to pattern catalyst-adsorbate techniques, that are central to heterogeneous catalysis. Identifying lively adsorption websites and steady adsorbate configurations is essential for understanding and designing catalysts, however it’s also fairly difficult as a result of advanced surface-molecular interactions. Traditional strategies, comparable to density practical concept (DFT) calculations and molecular dynamics simulations, are time-consuming and expensive, particularly for giant and sophisticated surfaces. DiG predicts adsorption websites and configurations, in addition to their chances, from the substrate and adsorbate descriptors. DiG can deal with varied kinds of adsorbates, comparable to single atoms or molecules being adsorbed onto several types of substrates, comparable to metals or alloys.
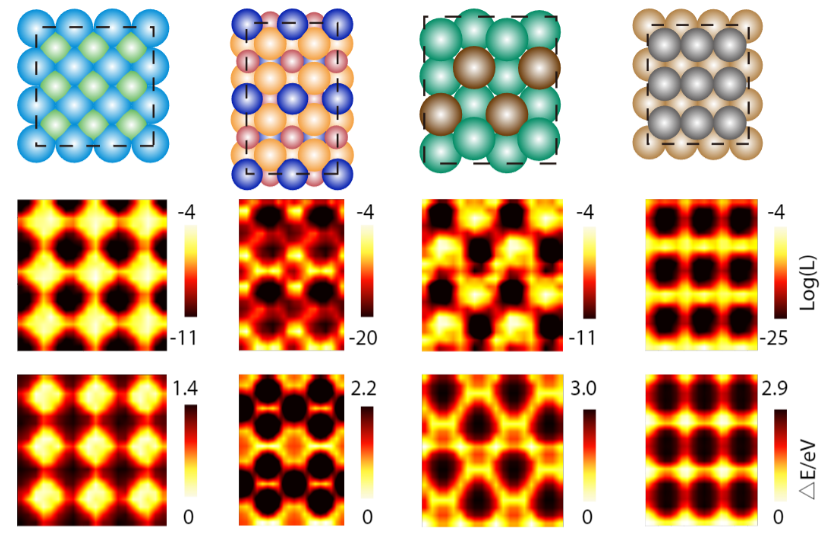
Applying DiG, we predicted the adsorption websites for quite a lot of catalyst-adsorbate techniques and in contrast these predicted chances with energies obtained from DFT calculations. We discovered that DiG may discover all of the steady adsorption websites and generate adsorbate configurations which might be much like the DFT outcomes with excessive effectivity and at a low price. DiG estimates the chances of various adsorption configurations, in good settlement with DFT energies.
Conclusion
In this weblog, we launched DiG, a deep studying framework that goals to foretell the distribution of molecular constructions. DiG is a big development from single construction prediction towards ensemble modeling with equilibrium distributions, setting a cornerstone for connecting microscopic constructions to macroscopic properties below deep studying frameworks.
DiG includes key ML improvements that result in expressive generative fashions, which have been proven to have the capability to pattern multimodal distribution inside a given class of molecules. We have demonstrated the flexibleness of this strategy on totally different lessons of molecules (together with proteins, and so forth.), and we’ve proven that particular person constructions generated on this method are chemically lifelike. Consequently, DiG permits the event of ML techniques that may pattern equilibrium distributions of molecules given applicable coaching information.
However, we acknowledge that significantly extra analysis is required to acquire environment friendly and dependable predictions of equilibrium distributions for arbitrary molecules. We hope that DiG evokes extra analysis and innovation on this course, and we look ahead to extra thrilling outcomes and affect from DiG and different associated strategies sooner or later.
[ad_2]
