[ad_1]
In a latest examine posted to the bioRxiv* preprint server, researchers study the phylogenetic relationships between Eulipotyphla coronaviruses (CoVs) and associated CoVs.
Study: Bioinformatic Analysis of the Spike Protein Cleavage Sites of Coronaviruses within the Mammalian Order Eulipotyphla. Image Credit: Cocoapowder / Shutterstock.com
Mammalian hosts for CoVs
Severe acute respiratory syndrome (SARS)-CoV-2 has considerably impacted international public well being. Horseshoe bats are the almost definitely pure reservoir for SARS-CoV-2.
Meanwhile, intermediate hosts have but to be definitively recognized and will embrace extremely diversified animals, similar to reptiles, avians, and mammals. Therefore, exploring small mammal CoVs and their zoonotic potential is important.
Members of the Eulipotyphla order, similar to shrews, hedgehogs, and moles, exhibit an in depth relationship with the Chiroptera order. Since the primary detection of CoVs in European hedgehogs in Germany, Erinaceus CoV (EriCoV) has been detected in Italy, France, and the United Kingdom. However, restricted research have targeted on Eulipotyphla mammals, which has left gaps in understanding these as small mammalian hosts for CoVs.
About the examine
In the current examine, researchers carry out bioinformatic analyses of CoVs for example evolutionary complexities and viral range throughout hosts. Spike protein sequences of various CoVs have been retrieved from the National Center for Biotechnology Information (NCBI) protein database. A most probability phylogenetic tree was constructed based mostly on spike sequences.
Study findings
The classification was obtained from the NCBI taxonomy browser below Orthocoronavirinae sub-family. EriCoVs from the identical species within the U.Okay., Germany, and Italy exhibited as much as 100% spike protein sequence id.
All unclassified Italian EriCoVs have been just like EriCoV isolates from the U.Okay. and Germany. Therefore, Italian EriCoVs might be categorized as Merbecovirus.
The two Amur hedgehog HKU31 CoVs from China fashioned a clade distinct from EriCoVs with as much as 79% sequence id. HKU31 and EriCoV spikes had a imply sequence id of 56.6% and 55.8%, with the Middle East respiratory syndrome (MERS)-CoV spike protein, respectively. Thus, hedgehogs may harbor different MERS-like CoVs and transmit them to different species.
Sequence identities inside Wencheng Sm shrew CoVs (WESVs) ranged from 90% to 99.74%, with WESVs forming a divergent group within the Alpha CoV genus. The highest spike sequence id between WESVs and different Alpha CoVs was 34.68% with HKU2, the Rhinolophus bat CoV. Likewise, shrew CoV emerged as a single clade in Alpha CoVs distinct from WESVs with a imply sequence id of 20% with different Alpha CoVs.
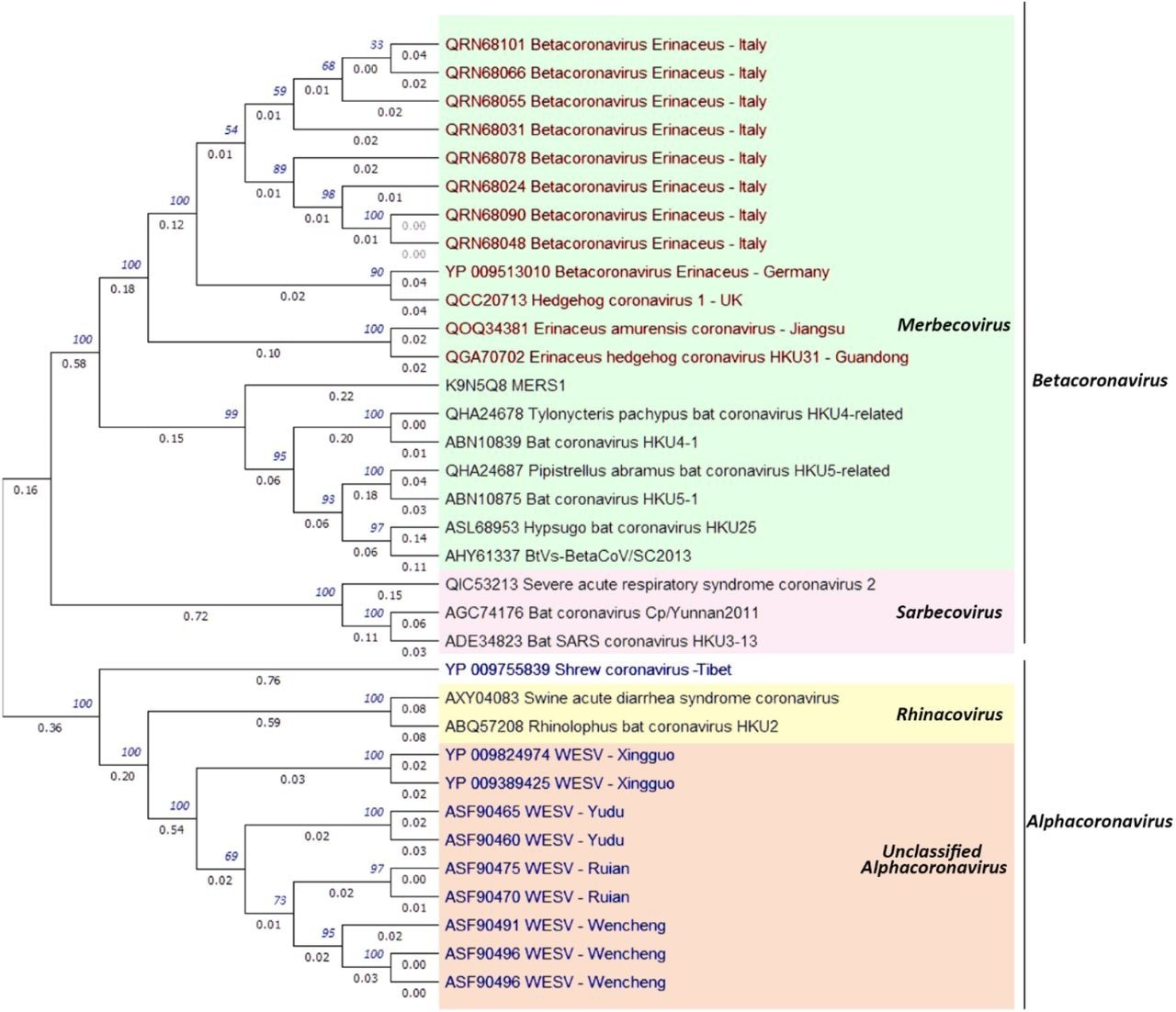
Maximum probability phylogenetic evaluation of the hedgehog coronavirus stains, shrew coronavirus strains, and associated alphacoronavirus and betacoronavirus strains based mostly on spike protein amino acid sequences. The isolates coloured in purple are hedgehog coronaviruses and the isolates coloured in blue are shrew coronaviruses.
The workforce analyzed 201 Alpha and Beta CoV strains for furin cleavage websites of their spikes. Cleavage websites in spikes have been recognized from 44 strains, 17 of which have bats as hosts, whereas 27 produce other mammalian hosts. Domestic animals, together with canines, cats, cattle, horses, and rabbits, have been additionally recognized as hosts for probably transmissible CoVs.
Additionally, furin sequences have been analyzed throughout animal species, together with mammals, hedgehogs, and shrews. Pairwise sequence alignment revealed a excessive diploma of conservation of furins throughout animals.
Furin cleavage websites in Eulipotyphla CoVs have been predicted by ProP and PiTou packages. PiTou revealed zero furin cleavage websites within the viral strains. Six WESVs have optimistic ProP predictions at atypical websites and is perhaps cleaved by different proteases or false positives.
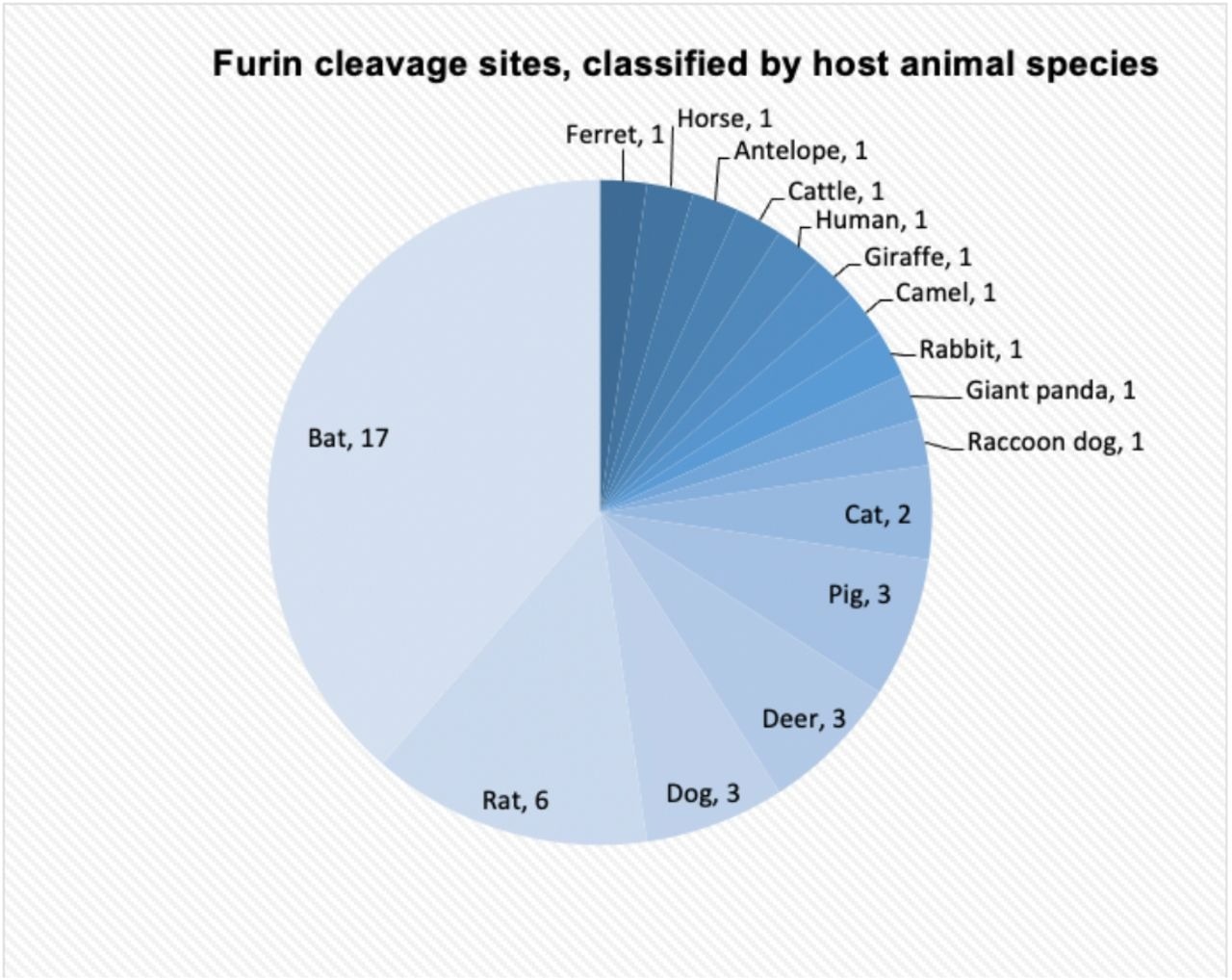
The variety of alphacoronaviruses and betacoronaviruses with furin cleavage websites equivalent to their host animals. Most viruses are hosted by bats (17 strains).
Two WESV spike proteins with excessive ProP consensus scores have been aligned and in comparison with the MERS-CoV spike. The putative proprotein convertase (PC) cleavage websites didn’t align with the spike cleavage website between S1 and S2 spike domains; as a substitute, these websites have been situated inside the S1 area.
A spike structural mannequin of 1 WESV was generated based mostly on the HKU2 spike, which has a excessive id with WESVs. The cleavage website within the mannequin was an uncovered loop accessible by proteases.
Conclusions
Shrew CoVs and WESVs have been discovered to diverge from a definite ancestor from different Alpha CoVs. Hedgehog CoVs together with HKU31 and EriCoV have been in the identical Merbecovirus sub-genus as MERS-CoV.
A PC cleavage website close to 512 was recognized in WESV, thus suggesting its zoonotic potential. PC5, PC7, and endoproteases, apart from furin, might cleave this website.
Animals harboring spike-cleaving furins, similar to bats, canines, cats, pigs, rats, horses, cattle, antelopes, and camels, have been extra more likely to host and transmit CoVs. Shrews and hedgehog CoVs could recombine with different animal CoVs and emerge as new viruses.
These information are important to grasp CoV evolution and transmission, as novel CoVs pose a big burden to international well being. Growing urbanization will increase animal-human interactions, with the commerce of small mammals in moist markets growing the probability of pathogenic spillover occasions. Therefore, broad animal surveillance ought to be carried out and people ought to take precautions to keep away from contact with these small mammals.
*Important discover
bioRxiv publishes preliminary scientific experiences that aren’t peer-reviewed and, due to this fact, shouldn’t be thought to be conclusive, information scientific follow/health-related conduct, or handled as established info.