[ad_1]
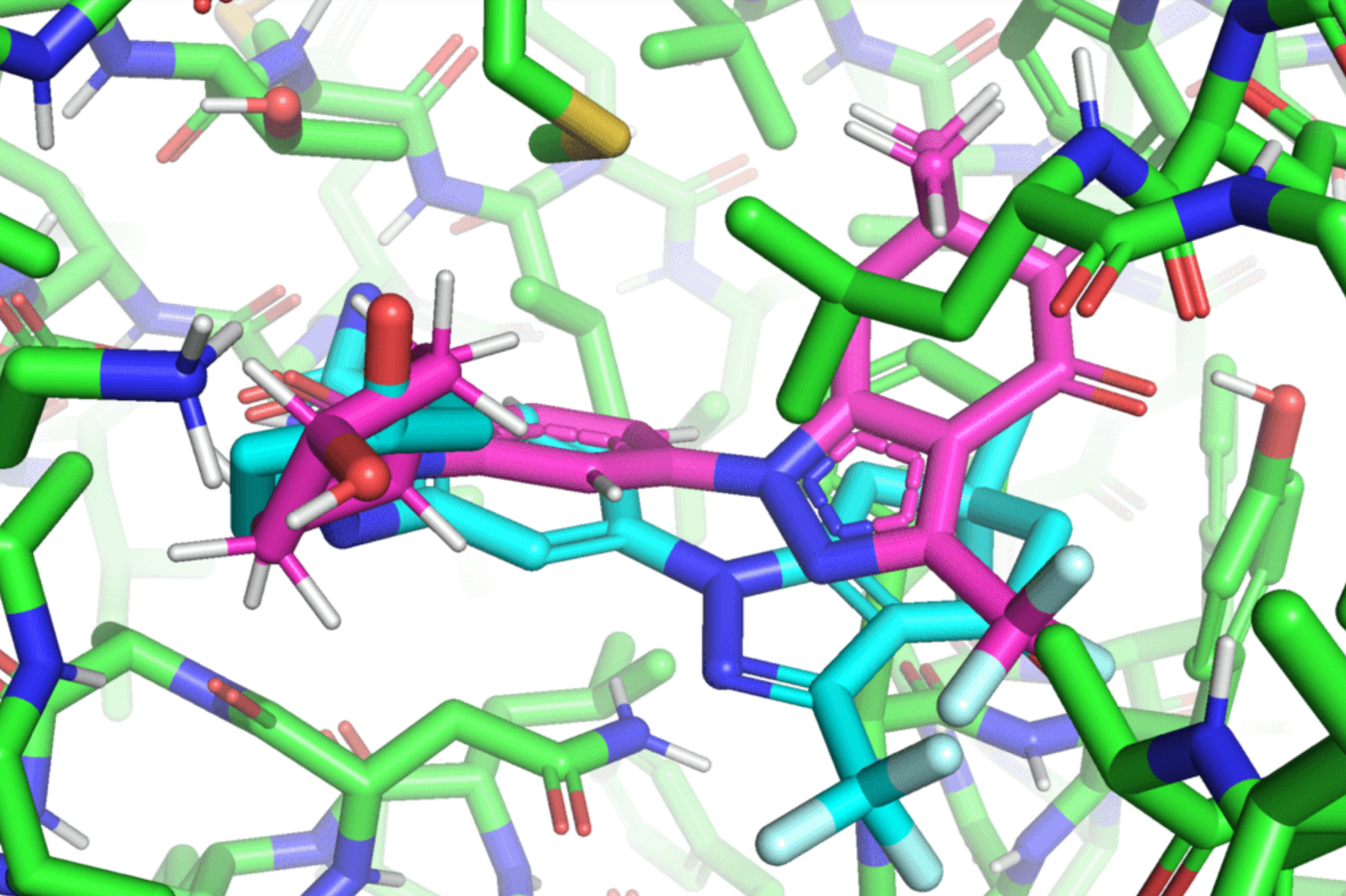
The entirety of the recognized universe is teeming with an infinite variety of molecules. But what fraction of those molecules have potential drug-like traits that can be utilized to develop life-saving drug therapies? Millions? Billions? Trillions? The reply: novemdecillion, or 1060. This gargantuan quantity prolongs the drug growth course of for fast-spreading illnesses like Covid-19 as a result of it’s far past what present drug design fashions can compute. To put it into perspective, the Milky Way has about 100 billion, or 1011, stars.
In a paper that can be introduced on the International Conference on Machine Learning (ICML), MIT researchers developed a geometrical deep-learning mannequin referred to as EquiBind that’s 1,200 occasions sooner than one of many quickest present computational molecular docking fashions, QuickVina2-W, in efficiently binding drug-like molecules to proteins. EquiBind relies on its predecessor, EquiDock, which makes a speciality of binding two proteins utilizing a way developed by the late Octavian-Eugen Ganea, a current MIT Computer Science and Artificial Intelligence Laboratory and Abdul Latif Jameel Clinic for Machine Learning in Health (Jameel Clinic) postdoc, who additionally co-authored the EquiBind paper.
Before drug growth may even happen, drug researchers should discover promising drug-like molecules that may bind or “dock” correctly onto sure protein targets in a course of generally known as drug discovery. After efficiently docking to the protein, the binding drug, also referred to as the ligand, can cease a protein from functioning. If this occurs to a necessary protein of a bacterium, it may kill the bacterium, conferring safety to the human physique.
However, the method of drug discovery will be expensive each financially and computationally, with billions of {dollars} poured into the method and over a decade of growth and testing earlier than last approval from the Food and Drug Administration. What’s extra, 90 p.c of all medicine fail as soon as they’re examined in people attributable to having no results or too many unintended effects. One of the methods drug corporations recoup the prices of those failures is by elevating the costs of the medicine which can be profitable.
The present computational course of for locating promising drug candidate molecules goes like this: most state-of-the-art computational fashions rely on heavy candidate sampling coupled with strategies like scoring, rating, and fine-tuning to get one of the best “fit” between the ligand and the protein.
Hannes Stärk, lead creator of the paper and a first-year graduate scholar suggested by Regina Barzilay and Tommi Jaakkola within the MIT Department of Electrical Engineering and Computer Science, likens typical ligand-to-protein binding methodologies to “trying to fit a key into a lock with a lot of keyholes.” Typical fashions time-consumingly rating every “fit” earlier than selecting one of the best one. In distinction, EquiBind instantly predicts the exact key location in a single step with out prior information of the protein’s goal pocket, which is named “blind docking.”
Unlike most fashions that require a number of makes an attempt to discover a favorable place for the ligand within the protein, EquiBind already has built-in geometric reasoning that helps the mannequin study the underlying physics of molecules and efficiently generalize to make higher predictions when encountering new, unseen knowledge.
The launch of those findings shortly attracted the eye of business professionals, together with Pat Walters, the chief knowledge officer for Relay Therapeutics. Walters recommended that the group attempt their mannequin on an already present drug and protein used for lung most cancers, leukemia, and gastrointestinal tumors. Whereas many of the conventional docking strategies didn’t efficiently bind the ligands that labored on these proteins, EquiBind succeeded.
“EquiBind provides a unique solution to the docking problem that incorporates both pose prediction and binding site identification,” Walters says. “This approach, which leverages information from thousands of publicly available crystal structures, has the potential to impact the field in new ways.”
“We were amazed that while all other methods got it completely wrong or only got one correct, EquiBind was able to put it into the correct pocket, so we were very happy to see the results for this,” Stärk says.
While EquiBind has acquired an excessive amount of suggestions from business professionals that has helped the group take into account sensible makes use of for the computational mannequin, Stärk hopes to seek out completely different views on the upcoming ICML in July.
“The feedback I’m most looking forward to is suggestions on how to further improve the model,” he says. “I want to discuss with those researchers … to tell them what I think can be the next steps and encourage them to go ahead and use the model for their own papers and for their own methods … we’ve had many researchers already reaching out and asking if we think the model could be useful for their problem.”
This work was funded, partly, by the Pharmaceutical Discovery and Synthesis consortium; the Jameel Clinic; the DTRA Discovery of Medical Countermeasures Against New and Emerging threats program; the DARPA Accelerated Molecular Discovery program; the MIT-Takeda Fellowship; and the NSF Expeditions grant Collaborative Research: Understanding the World Through Code.
This work is devoted to the reminiscence of Octavian-Eugen Ganea, who made essential contributions to geometric machine studying analysis and generously mentored many college students — a superb scholar with a humble soul.
