[ad_1]

The mixture of the setting a person experiences and their genetic predispositions determines the vast majority of their danger for varied illnesses. Large nationwide efforts, corresponding to the UK Biobank, have created massive, public sources to higher perceive the hyperlinks between setting, genetics, and illness. This has the potential to assist people higher perceive find out how to keep wholesome, clinicians to deal with diseases, and scientists to develop new medicines.
One problem on this course of is how we make sense of the huge quantity of scientific measurements — the UK Biobank has many petabytes of imaging, metabolic assessments, and medical information spanning 500,000 people. To greatest use this information, we want to have the ability to characterize the knowledge current as succinct, informative labels about significant illnesses and traits, a course of known as phenotyping. That is the place we will use the power of ML fashions to choose up on delicate intricate patterns in massive quantities of knowledge.
We’ve beforehand demonstrated the power to make use of ML fashions to rapidly phenotype at scale for retinal illnesses. Nonetheless, these fashions have been skilled utilizing labels from clinician judgment, and entry to clinical-grade labels is a limiting issue as a result of time and expense wanted to create them.
In “Inference of chronic obstructive pulmonary disease with deep learning on raw spirograms identifies new genetic loci and improves risk models”, printed in Nature Genetics, we’re excited to focus on a technique for coaching correct ML fashions for genetic discovery of illnesses, even when utilizing noisy and unreliable labels. We display the power to coach ML fashions that may phenotype instantly from uncooked scientific measurement and unreliable medical report data. This decreased reliance on medical area consultants for labeling enormously expands the vary of functions for our approach to a panoply of illnesses and has the potential to enhance their prevention, prognosis, and therapy. We showcase this technique with ML fashions that may higher characterize lung perform and continual obstructive pulmonary illness (COPD). Additionally, we present the usefulness of those fashions by demonstrating a greater capacity to determine genetic variants related to COPD, improved understanding of the biology behind the illness, and profitable prediction of outcomes related to COPD.
ML for deeper understanding of exhalation
For this demonstration, we centered on COPD, the third main explanation for worldwide dying in 2019, during which airway irritation and impeded airflow can progressively scale back lung perform. Lung perform for COPD and different illnesses is measured by recording a person’s exhalation quantity over time (the report is named a spirogram; see an instance under). Although there are pointers (known as GOLD) for figuring out COPD standing from exhalation, these use just a few, particular information factors within the curve and apply mounted thresholds to these values. Much of the wealthy information from these spirograms is discarded on this evaluation of lung perform.
We reasoned that ML fashions skilled to categorise spirograms would be capable to use the wealthy information current extra utterly and end in extra correct and complete measures of lung perform and illness, much like what we now have seen in different classification duties like mammography or histology. We skilled ML fashions to foretell whether or not a person has COPD utilizing the complete spirograms as inputs.
The widespread technique of coaching fashions for this downside, supervised studying, requires samples to be related to labels. Determining these labels can require the hassle of very time-constrained consultants. For this work, to indicate that we don’t essentially want medically graded labels, we determined to make use of quite a lot of broadly out there sources of medical report data to create these labels with out medical professional assessment. These labels are much less dependable and noisy for 2 causes. First, there are gaps within the medical information of people as a result of they use a number of well being providers. Second, COPD is commonly undiagnosed, that means many with the illness is not going to be labeled as having it even when we compile the whole medical information. Nonetheless, we skilled a mannequin to foretell these noisy labels from the spirogram curves and deal with the mannequin predictions as a quantitative COPD legal responsibility or danger rating.
Predicting COPD outcomes
We then investigated whether or not the chance scores produced by our mannequin may higher predict quite a lot of binary COPD outcomes (for instance, a person’s COPD standing, whether or not they have been hospitalized for COPD or died from it). For comparability, we benchmarked the mannequin relative to expert-defined measurements required to diagnose COPD, particularly FEV1/FVC, which compares particular factors on the spirogram curve with a easy mathematical ratio. We noticed an enchancment within the capacity to foretell these outcomes as seen within the precision-recall curves under.
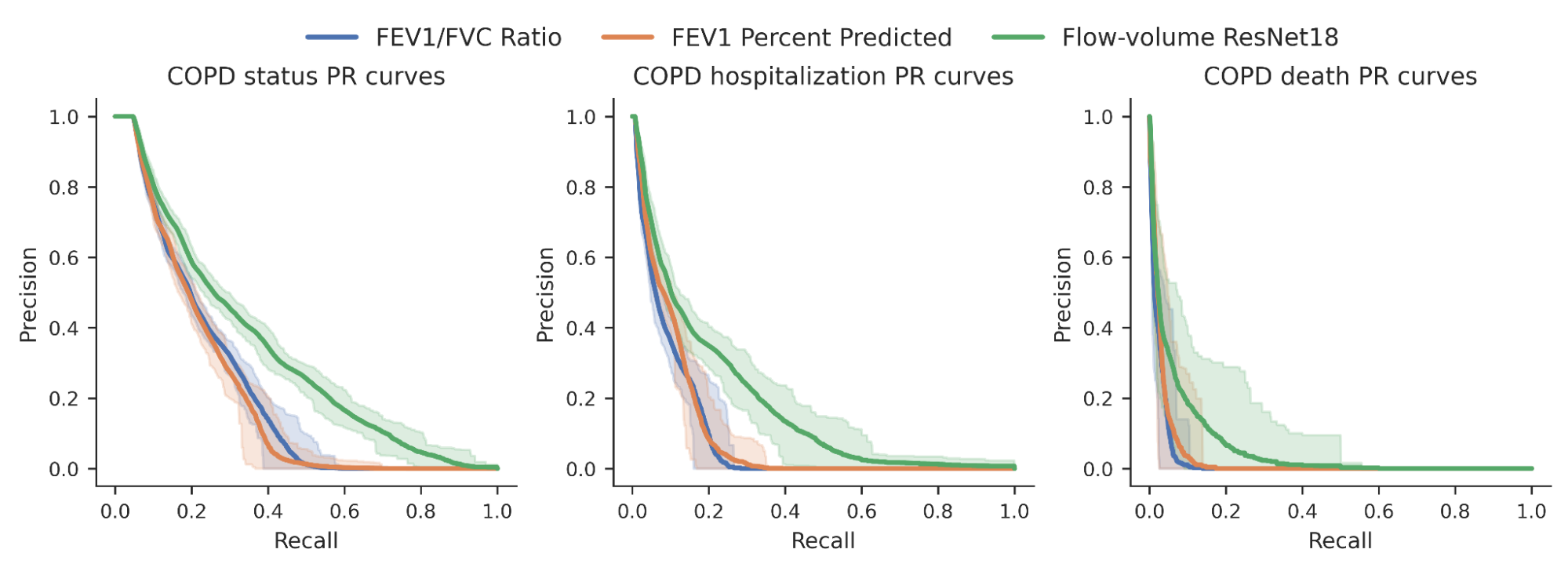 |
| Precision-recall curves for COPD standing and outcomes for our ML mannequin (inexperienced) in comparison with conventional measures. Confidence intervals are proven by lighter shading. |
We additionally noticed that separating populations by their COPD mannequin rating was predictive of all-cause mortality. This plot means that people with increased COPD danger usually tend to die earlier from any causes and the chance in all probability has implications past simply COPD.
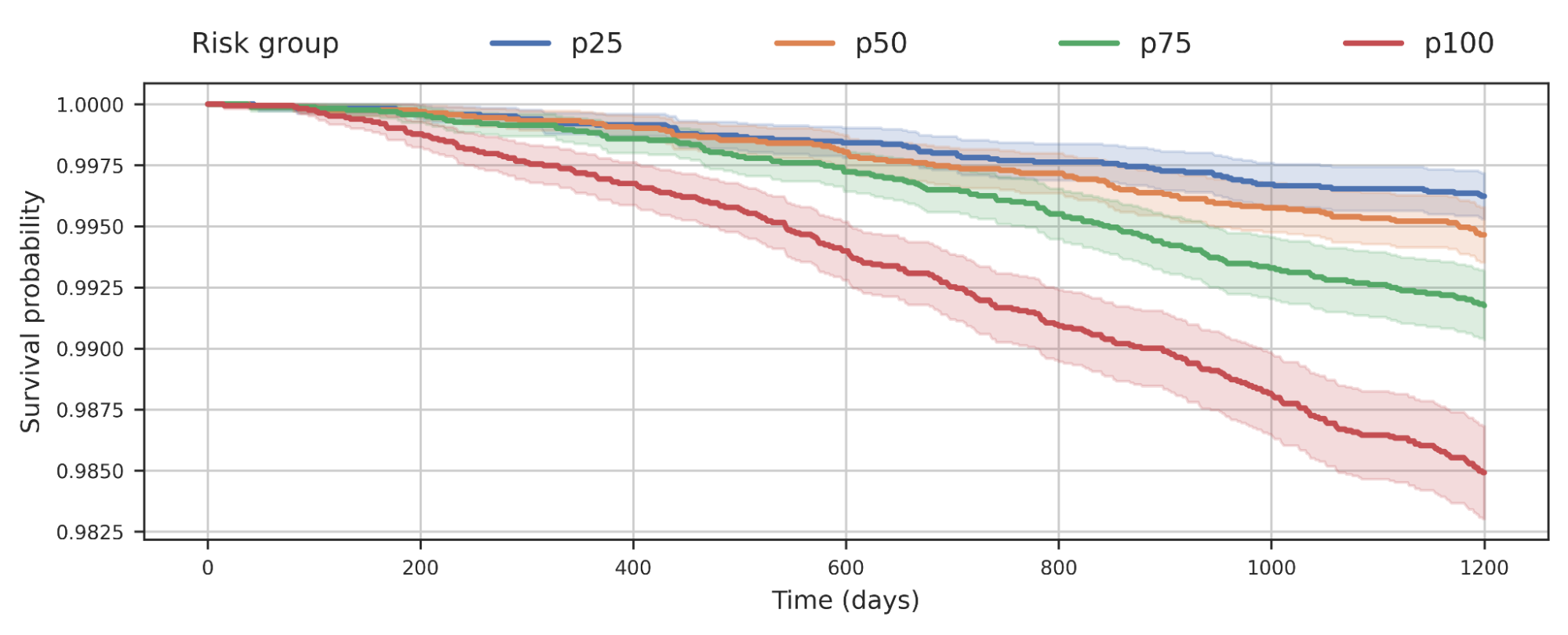 |
| Survival evaluation of a cohort of UK Biobank people stratified by their COPD mannequin’s predicted danger quartile. The lower of the curve signifies people within the cohort dying over time. For instance, p100 represents the 25% of the cohort with biggest predicted danger, whereas p50 represents the 2nd quartile. |
Identifying the genetic hyperlinks with COPD
Since the objective of huge scale biobanks is to deliver collectively massive quantities of each phenotype and genetic information, we additionally carried out a check known as a genome-wide affiliation examine (GWAS) to determine the genetic hyperlinks with COPD and genetic predisposition. A GWAS measures the energy of the statistical affiliation between a given genetic variant — a change in a selected place of DNA — and the observations (e.g., COPD) throughout a cohort of circumstances and controls. Genetic associations found on this method can inform drug improvement that modifies the exercise or merchandise of a gene, in addition to broaden our understanding of the biology for a illness.
We confirmed with our ML-phenotyping technique that not solely can we rediscover nearly all identified COPD variants discovered by guide phenotyping, however we additionally discover many novel genetic variants considerably related to COPD. In addition, we see good settlement on the impact sizes for the variants found by each our ML strategy and the guide one (R2=0.93), which gives sturdy proof for validity of the newly discovered variants.
Finally, our collaborators at Harvard Medical School and Brigham and Women’s Hospital additional examined the plausibility of those findings by offering insights into the doable organic function of the novel variants in improvement and development of COPD (you may see extra dialogue on these insights within the paper).
Conclusion
We demonstrated that our earlier strategies for phenotyping with ML may be expanded to a variety of illnesses and might present novel and useful insights. We made two key observations through the use of this to foretell COPD from spirograms and discovering new genetic insights. First, area data was not essential to make predictions from uncooked medical information. Interestingly, we confirmed the uncooked medical information might be underutilized and the ML mannequin can discover patterns in it that aren’t captured by expert-defined measurements. Second, we don’t want medically graded labels; as an alternative, noisy labels outlined from broadly out there medical information can be utilized to generate clinically predictive and genetically informative danger scores. We hope that this work will broadly broaden the power of the sector to make use of noisy labels and can enhance our collective understanding of lung perform and illness.
Acknowledgments
This work is the mixed output of a number of contributors and establishments. We thank all contributors: Justin Cosentino, Babak Alipanahi, Zachary R. McCaw, Cory Y. McLean, Farhad Hormozdiari (Google), Davin Hill (Northeastern University), Tae-Hwi Schwantes-An and Dongbing Lai (Indiana University), Brian D. Hobbs and Michael H. Cho (Brigham and Women’s Hospital, and Harvard Medical School). We additionally thank Ted Yun and Nick Furlotte for reviewing the manuscript, Greg Corrado and Shravya Shetty for assist, and Howard Yang, Kavita Kulkarni, and Tammi Huynh for serving to with publication logistics.



